-
Notifications
You must be signed in to change notification settings - Fork 875
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Lables of checkerboard plot in example of mcnemar_table do not match the documentation #987
Comments
|
@rasbt: I just wanted to chime in and confirm @ftnext's findings here (as well as those on his other issue, i.e., #988). The documentation is inconsistent in terms of which side of the table are labeled as model1 vs. model2 and correct vs. incorrect. Based on the current implementation, the documentation examples generally do not reflect things accurately given the current mlxtend code (as of Dec 2024). Specifically, for the following code: import numpy as np
from mlxtend.evaluate import mcnemar_table
# The correct target (class) labels
y_target = np.array([0, 0, 0, 0, 0, 1, 1, 1, 1, 1])
# Class labels predicted by model 1
y_model1 = np.array([0, 1, 0, 0, 0, 1, 1, 0, 0, 0])
# Class labels predicted by model 2
y_model2 = np.array([0, 0, 1, 1, 0, 1, 1, 0, 0, 0])
tb = mcnemar_table(y_target=y_target,
y_model1=y_model1,
y_model2=y_model2)
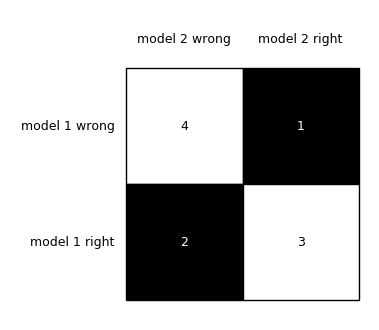
print(tb)The documentation claims output of: [[4 1]
[2 3]]When in reality you get: [[4 2]
[1 3]]In reality, the truth for this example ought to be: # both correct: 4
# both incorrect: 3
# only m1 correct: 2
# only m2 correct: 1
# m2c m2i
# m1c 4 2
# m1i 1 3I believe these issues cropped up with some older revisions, i.e., #664 and #744. Thankfully, none of this affects test results, but the following documentation pages need to be cleaned up since they lead to confusion:
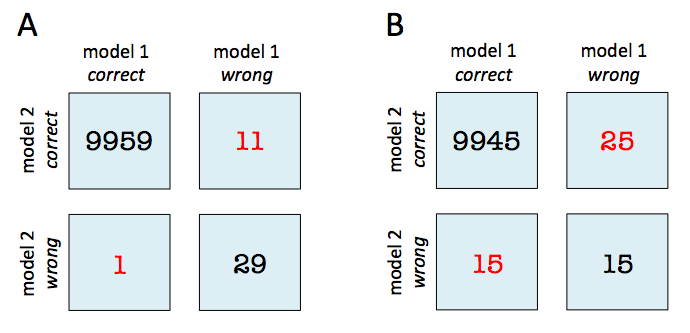
For the examples in the documentation to be correct, all tables should follow this format (as in the first example of the second link above): However, notice that the subsequent examples use the wrong labels: Finally, as @ftnext points out, this example is simply wrong in terms of its labels compared to the ground truth (i.e., if the figure below were correct, the bottom right corner ought to be 4 -- the labels here really should be reversed): |
|
Thanks for the note, I appreciate it. I am currently out with an injury but will bookmark this and revisit it in the upcoming weeks. |
Wishing you a quick recovery! |
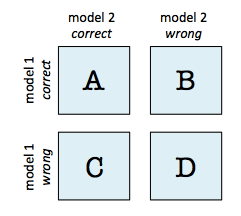
Describe the documentation issue
https://rasbt.github.io/mlxtend/user_guide/evaluate/mcnemar_table/#example-2-2x2-contingency-table
The labels of the checkerboard plot do not seem to match the description of the returned value
tbisfrom returns in https://rasbt.github.io/mlxtend/user_guide/evaluate/mcnemar_table/#api
tb[0, 0], a) is # of samples that model 1 and 2 got righttb[0, 1], b) is # of samples that model 1 got right and model 2 got wrongtb[1, 0], c) is # of samples that model 2 got right and model 1 got wrongtb[1, 1], d) is # of samples that model 1 and 2 got wrongSuggest a potential improvement or addition
I think that reversing each labels will work.
The text was updated successfully, but these errors were encountered: